|
|
|
||||||||||||||||||||
4.1. Survey of genetic elements introduced into approved transgenic cropsThe structural genes and the respective regulatory sequences (promoters and terminators) which have been used as transgenes are summarised in the following sections. Depending on their frequency of use in approved genetically modified crops and the probability of a 'natural' occurrence of these sequences in food, sequences within single genetic elements are applicable for the screening for GMO-food. Genetic elements that have been used in isolated cases may allow specific detection of the given product for as long as the respective element is not employed in further approved products. Thus, methods based on the detection of sequences within a single genetic element will in the long run be better suited for screening purposes. 4.1.1 Survey of the structural genes usedAlmost 35 distinct structural genes (including variants) have been used for the generation of the currently approved transgenic crops (Structural genes introduced into approved transgenic crops). Some of these genes such as accd, accS, sam-k and some genes coding viral coat proteins occur only in a single genetically engineered product. Therefore, the identification of sequences of one of these genes in food would represent a product-specific detection method provided the actual sample did not contain the natural sources of these sequences (e.g. from bacteriophages or plant viruses). The most frequently used transgene is the nptII gene, originally isolated from the bacterial transposon 5. The nptII gene has been introduced into 17 out of the 28 approved agricultural crops. In 16 products it functioned as a marker gene under the control of a eucaryotic promoter; thus, nptII sequences seem to be well suited for screening purposes. It should be noted, however, that nptII occurs frequently in bacteria found in the environment (Smalla et al., 1993; Redenbaugh et al., 1994). The presence of these naturally occurring bacteria in a sample may, therefore, lead to a false-positive result. Future transgenic crops are expected to contain fewer or no marker genes in the final products since marker-free insertion techniques or methods to eliminate marker genes from transgenic plants (for review see Yolder and Goldbrough, 1994 and references in Niederhauser et al., 1996) and microorganisms (Sanchis et al., 1997) are already available. Other structural genes have been employed less frequently. From the 28 approved crops, variants of the -endotoxin gene from Bacillus thuringiensis or of the bar gene originally isolated from Streptomyces hygroscopicus are found in 6 products each. Variants of the CP4 epsps gene from Agrobacterium, the -lactamase gene and of the polygalacturonase gene have been introduced in 3 to 5 products each (Figure 4). In these cases various factors may have to be assessed to judge the applicability of DNA-based detection methods: (i) the presence of the transgene in the respective transformation event (line); (ii) the 'completeness' of the respective sequence (incompletely transferred, or 'truncated' or 'altered' versions of genes may be present); (iii) the use of 'synthetic' versions of genes that have an altered codon usage in order to optimise gene expression in the host organism. 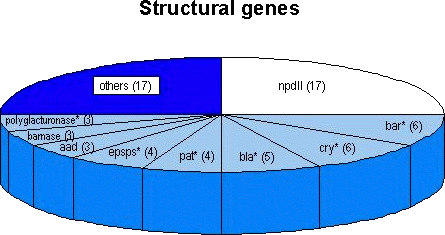
Figure 4: Number of occurrences of the most frequently used transgenes introduced into the currently approved genetically engineered agricultural crops (in total 28 distinct products were approved; see text). In some cases (indicated by asterisks) distinct variants of genes or 'synthetic' versions were used. (See also Structural genes introduced into approved transgenic crops.) Both the gene encoding for barnase and the aad gene are present in three products each, whereas a synthetic version of the pat gene can be found in four products. Seventeen other genes are present in one or two of 17 different products. Sequences from structural genes originating from homologous sources (up to now mostly antisense constructs in tomatoes) are suitable for the detection of GMOs only if certain prerequisites are fulfilled. When the coding sequences occur in the (copies of the) transgene and also in the naturally occurring copies of the gene, a clever choice of primers for a PCR assay may allow discrimination of the amplification products of the native gene and the transgene by the length of the amplified fragment. This can be achieved if the two primers bind to sequences on the chromosomal gene that are situated on different (normally adjacent) exons. Whereas analysis of conventional products would result in the amplification of a single long fragment that includes the sequence of the intron between the respective primer binding sites, analysis of the corresponding genetically modified product would result in the appearance of an additional, shorter amplification product lacking the intron sequence (since transgenes originate from c-DNA sequences). A description of this methodology was contained in the petition for the genetically engineered tomato from Zeneca (Petition from Zeneca for genetically modified tomatoes, 1995). However, such a strategy requires not only the knowledge of the c-DNA sequence (of the transgene) but also precise information about the intron-exon boundaries of the chromosomal gene, which is not always available.
|
|||||||||||||||||||||




 antibodies on hand
antibodies on hand 