|
|
|
||||||||||||||||||||
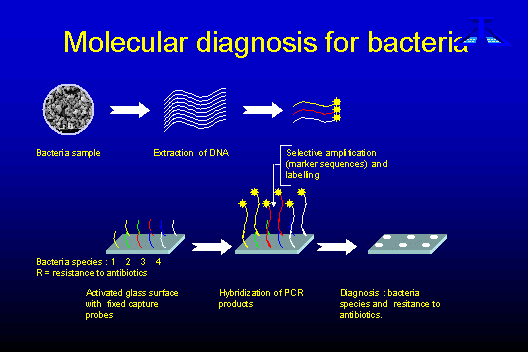
Biochips: A powerful tool for multiple and fast analysis of genes and DNA sequencesBiochip technology combines two in one: multiparametric molecular analysis and quick identification of genomic material in biological samples. In this article José Remacle and Stéphanie Warnon from the University of Namur (FUNDP, Laboratoire de Biochimie Cellulaire, Belgium) introduce this promising technology, show its possible applications and report on current research activities. General introduction to biochipsDNA microarrays or biochips are formed by grafting multiple capture probes onto a surface. The word "biochip" derives from the computer term "chip". Although silicon surfaces bearing printed circuits can be used for DNA binding, the term biochip is now broadly used to describe all surfaces bearing microscopic spots, each one being formed by specific capture probes. The capture probes are chosen to complement the target sequence to be detected. Each capture probe will bind to its corresponding target sequence. The purpose of the chips is to detect many genes present in a sample in one assay rather than performing individual gene assays as is the practice e.g. in so-called multiwells, plates with 96 wells, where the reactions take place. The huge amount of information coming from the genome sequence and other research genome programs cannot be utilised to the full without the availability of methods such as biochips which enable these genes or specific DNA sequences to be detected in biological samples. DNA microarray technology is an example of the enormous efforts undertaken in the genomic field in the last few years Many applications and technologies adapted to these applications use biochips. One of the most common applications is in the determination of gene expression for a given tissue. This is useful in correlating a change in gene expression with pathology or with cell response to drugs. These applications have been well developed by companies like Affymetrix or Incyte using high density chips bearing either small or long capture probes. Researchers and pharmaceutical companies chips searching for new drug targets have displayed most interest in the potential of such chips. Chips can also be utilised in the determination of Single Nucleotide Polymorphism (SNP) allowing one to discriminate between sequences differing by a single nucleotide. Using small capture probes (Affymetrix) and changing the stringency of hybridisation using electric repulsion (Nanogen) we have succeeded in doing this. However the test is far from being quantitative and sensitivity is very poor. The determination of SNP is of relevance when studying genetic mutations which can be linked to genetic disease or in differentiating close species or homologue genes. The proposed strategy for biochipsWe are focusing our efforts on the developement of biochips as tools for every day diagnosis so that large numbers of genes or sequences can be tested in one assay giving the doctor all the information s/he needs to treat the patient. In practice biochips for the detection of Staphyloccocus first allows the identification of Staphylococcus as the source of infection. Secondly they enable us to establish whether the Staphylococcus are resistant to methicilin thereby helping to determine the appropriate antibiotic treatment. The chips used are precisely defined products consisting of a fixed and limited number of carefully chosen capture probes. The choice of the DNA sequences to be detected is made according to the answer expected. The chips give the user the information required but no more. In this way the size of the biochip is maintained at a minimum and product costs are drastically reduced. Our experience in the development of quantitative assays for DNA and fixation were to our advantage in the optimisation of the DNA probe hybridisation thereby obtaining assays with good sensitivity and reproducibility. Indeed, the main problem encountered in chip technology is the following: with miniaturisation of the detection spots, there is a concomitant lowering of the detection signal and an increase in noise so that the assay sensitivity is also diminished. The proposed chip technologyA general presentation of the chips is given in figure 1:
Fixation of the DNA capture probesThe capture probes are covalently linked by their 5' end in order to control both the amount fixed and the length of the sequence available for hybridisation. The capture probes are single strand and specially designed for high yield hybridisation. The laboratory know-how involved here is very important when seeking to obtain high sensitivity. Spotting
DetectionDetection is commonly performed by fluorescence after incorporation of one fluorochrome in the target sequences during duplication or amplification. A double labelling system is useful when comparing two samples. However, the fluorescent scanner is not an easy machine to use and is too expensive for clinical laboratories. We sought to develop a detection method which could be adapted for routine laboratory use. We developed a new process which permits a darkening of the spots testing positive for DNA binding. These spots can be analysed with a colorimetric scanner with a resolution of 10 micrometers which is sufficient to quantify the spots. Analysis software which recognises and identifies the spots was developed. It also quantifies the labelling and perfoms a statistical analysis of the data. The scanner is available together with the software and the computer. This technology is covered by a filed patent (EU 99870106.4). The main advantage of this detection system is its simplicity and very low cost. The EU Project"New technology in food sciences encounter a multiplicity of recently released gmo's" (gmo chips)" This year sees our participation in a new gmo chips project with six other partners. The project aims to develop biochips capable of detecting genetically modified organisms (gmo's) in food. Reliable detection methods are essential for the labelling of foodstuffs containing gmo's and for EU border controls. Over the last years, there has been a dramatic and continuing increase of the surface area planted with transgenic crops. The five principal transgenic foodstuffs are maize, soybean, rapeseed / canola, tomato and potato. The European Union informs the consumers of the presence of a transgenic foodstuff by labelling of "substantially equivalent" ingredients (Food Regulation 258/97 and the Council regulations 1139/98, 49/2000 and 50/2000). The fortuitous presence of recombinant DNA or modified protein above a defined threshold of 1 % of ingredients in the foods will lead to unambiguous food labelling. The development and application of reliable and quantitative analytical detection methods are thus essential in order to perform and to control food labelling as well as for the possible development of "gmo free" production schemes and to control plant importation at the EU border. Biochip technology combines the advantages of clear identification and a multiparametric approach to detect unexpected gmo's via their specific patterns. Further information:
|
|||||||||||||||||||||



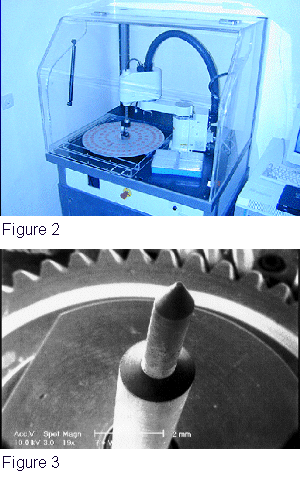 We developed
a specially devised robot to lay the capture probes on a glass slide.
The robot was developed in collaboration with
We developed
a specially devised robot to lay the capture probes on a glass slide.
The robot was developed in collaboration with

 antibodies on hand
antibodies on hand 