|
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3 State of the Art Genetic Engineering for Plant Protection3.1 Resistance of Transgenic PlantsStrategies using genetic engineering to achieve disease and pest resistant plants are advancing rapidly. Different degrees of resistance against insects, viruses, fungi and bacteria have been reached with various crop species. Table 3.1 summarizes the principal approaches and the state of development. The following chapters describe these approaches in more detail. Safety aspects of these approaches are discussed in chapter 4.1. 3.1.1 Resistance to Plant-Feeding InsectsWorld-wide losses resulting from plant-feeding insects are still more than 10 % of total production, despite the intensive use of a large number of chemical insecticides.[7] The use of these agrochemicals is not only an important economical factor in agriculture, with annual expenses of more than $7.8 billion world-wide, but has also often resulted in ecological problems.[1] In addition, more than 500 target insect species have developed resistance to chemical insecticides.[8] For these reasons, several approaches of genetically engineered insect resistance have been developed. The use of Bacillus thuringiensis toxins has by far been the most widely used and successful strategy, and several Bt expessing crops have obtained marketing clearance (see chapter 4.4). Some other approaches for insect resistance in transgenic plants are in development and may lead to successful insect resistance in the near future. 3.1.1.1 Bacillus Thuringiensis δ-EndotoxinsBiological Basis Bacillus thuringiensis (Bt) is a common soil bacterium which forms resistant resting structures under adverse conditions, so called spores. During formation of these spores, Bt produces a crystal which is predominantly comprised of one or more proteins called δ-endotoxins, Bt toxins or insecticidal crystal proteins. These δ-endotoxins possess insecticidal activity when ingested by certain insects. Intensive investigations have led to detailed knowledge of the mechanism and specificity of Bt toxin activity. Upon ingestion by susceptible insect larvae, the crystalline inclusions are solubilized in the midgut, releasing one or more proteins of different sizes. Rapid proteolytic cleavage of these polypeptides then produces the active toxic proteins. These activated toxins bind to specific high-affinity receptors on the insect midgut cell membranes, leading to the formation of pores that disturb the cellular osmotic balance. Within minutes, midgut cells are paralyzed and disrupted. [9] Bacillus thuringiensis bacteria have been used since 1938 to produce an insecticidal spray against certain insect pests. These products were, and still are, produced by fermentation of single Bt strains in crude, inexpensive media. Numerous strains of Bt are currently known. Each strain produces differing numbers of δ-endotoxins with various insecticidal activities. A given δ-endotoxin is typically insecticidal only towards a narrow spectrum of insect targets. Screening efforts have yielded strains insecticidal to Lepidoptera, Diptera, Coleoptera, and even some strains effective against nematodes. Although Bt bacteria are an effective microbial pesticide, a number of biological constraints limit their use: The Bt δ-endotoxins are short lived when sprayed on crops, thereby reducing the residual activity and necessitating many applications during a growing season. Bt, as with other spray-on insecticides, is difficult to deliver to insect species which burrow into their host plant, hide under leaves or live primarily under the soil surface[8]
Table 3.1: Strategies used to archieve pest or disease resistant transgenic plants (see text for sources). These limitations can be overcome by expressing Bt toxins in transgenic plants. Since Bt toxin is the product of a single gene, and its safe use and efficacy has a long history, the δ-endotoxin genes were among the first genes of commercial interest to be engineered into plants. In the beginning, plants were transformed with full length Bt toxin genes. These plants mostly showed only low levels of toxin mRNA and protein and did not exert significant insecticidal activity. Better results were obtained using truncated versions of Bt genes, but in many cases the level of Bt toxin expression and therefore protection was still to low to be of agronomic relevance.[8] Significant increases were achieved by modification of the truncated structural gene that had either none or only a minor effect on the encoded amino acid sequence.[9] Alterations affected 5'-or 3'- terminal regulatory mRNA sequences, mRNA secondary structure, G+C content and/or codon usage. For example, Koziel et al. (1993) synthesized a truncated crylA(b) gene encoding amino acids 1-648 of crylA(b) gene from Bacillus thuringiensis var. kurstaki HD-1. This synthetic gene was completely redesigned to replace the bacterial codons with maize- preferred codons. The synthetic gene was combined with different promoters and brought into maize. Field trials showed that the transgenic maize lines provided season-long protection from repeated heavy infestations of European corn borer (Ostrinia nubilalis, Lepidoptera), which totally devastated control plants.[10] Although the redesign of the δ-endotoxin genes is a successful means to enhance the Bt toxin production in plants, it suffers from the disadvantage of being laborious and expensive. In a recently published paper, a promising new solution for enhanced translation efficiency of Bt toxin genes was reported.[11] In this work, the transgene was not introduced into the nucleus, but into plastids, a group of small organelles of plant cells including chloroplasts with an own genome called the plastone. The plastids are thought to be derived from endosymbiontic bacteria and therefore have a transcriptional and translational machinery of procaryotic origin. An unmodified Bt toxin gene was incorporated into the tobacco plastid genome by plastid transformation. Since a plant cell may contain up to 50'000 copies of a plastid, the δ-endotoxin gene was present in several thousands of copies per cell in transgenic plants. The plastid transformation resulted in an accumulation of an unprecedented 3-5 % of soluble protein of tobacco cells as Bt toxin and an extremely high toxicity to the insects Heliothis virescens, Helicoverpa zea and Spodoptera exigua. Apart from the high expression level of unaltered Bt genes, the new method of plastid transformation has the additional advantages that position effects can be avoided because the site of introduction of the transgene can be targeted and the transgenes contained in the plastids are not transmitted to other plants by pollen.[11] State of Development As already mentioned, transgenic maize, potato and cotton expressing Bt toxins are among the first transgenic plants arriving on the market in the USA. During the last years, hundreds of field trials in several countries have proven the efficacy of this approach in controlling important insect pests. Table 3.2 indicates field-tested crops expressing Bt toxins. At present, the majority of transgenic crops containing Bt genes have been transformed with cryI genes which have activity against the lepidopterans, including the tobacco hornworm, tobacco budworm, tomato pinworm, corn earworm and European corn borer. More recently, the cryIIIA gene has been engineered into plants for protection against coleopterans, including the Colorado potato beetle. Bt δ-endotoxin genes with activity against insects of the genus Diabrotica, such as corn rootworm, a major maize pest, will likely be produced in the next generation of transgenic plants.[8]
Table 3.2: Field tested crops expressing Bt toxins in the European Union and the USA. Compiled from [12] and [13] 3.1.1.2 Inhibition of Insect Digestive EnzymesBiological Basis Many plant species protect themselves against plant-feeding insects by producing proteins which interfere with insect digestion. Proteases for example are enzymes which brake down proteins in the insect digestive system. Protease inhibitors block the action of these proteases. Most plant protease inhibitors do not effect endogenous plant proteases, but specifically inhibit animal and microbial protease enzymes. This suggests they may be involved in the protection of vulnerable plant tissues from pest and pathogen attack by antinutritional interaction with digestive enzymes. Protease inhibitors are widely distributed within the plant kingdom and can accumulate to particularly high levels in seeds and storage organs. The detrimental effect of protease inhibitors was proven by feeding insects an artificial diet. Rapid systemic accumulation of protease inhibitor mRNA and _proteins was demonstrated in tomato and potato in response to insect attack or mechanical wounding.[9] Another enzyme interfering with insect digestion is α-amylase inhibitor (αAI). This inhibitor blocks the action of α-amylase, an enzyme used by insects to degrade starch.[14] State of Development The first protease inhibitor protein expressed in transgenic plants was the cowpea trypsin inhibitor (CpTI). A full length CpTI DNA sequence was brought under control of the CaMV 35S promoter and transferred into tobacco. Bioassays considering insect biomass and survival and percentage of leaf area have conclusively established that CpTI-expressing plants have significant resistance to Heliothis virescens and to a broad range of lepidopteran pests feeding on tobacco. Recent field trials with transgenic tobacco expressing CpTI demonstrated that CpTI had a negative effect on both larval survival and plant damage, but the effect was not always significant. The tomato protease inhibitors PH and the potato proteinase inhibitor PPI II represent two other proteinase inhibitors which have been introduced into plants. They adversely affected some insect species like Manduca sexta (tobacco hornworm, lepidoptera) and Chrysodeixis eriosoma (green looper, lepidoptera). In contrast, no inhibition of feeding was achieved with transgenic tobacco producing PI-I inhibitor from tomato or soybean Kunitz trypsin inhibitor (SBTI), indicating that not every protease inhibitor can be employed for control of a specific pest species. [15) Two recent developments made possible the breeding of peas resistant to bruchid beetles, important pests of seeds in post harvest storage, by using α-amylase inhibitor ( αAI). First, it was shown that the growth of two bruchid species is inhibited by low doses of bean αAI, and second, a system for genetic transformation of garden pea became available. The coding sequence of the bean αAI-gene was attached to a seed-specific promoter and the chimeric gene was introduced into garden peas (Pisum sativum L). The seeds of the resulting transgenic peas showed a high degree of resistance to the bruchids cowpea weevil and Azuki bean weevil. This achievement may be particularly important in developing countries, not only because of the large amount of post-harvest losses, but also because of the importance of legumes such as peas as a source of protein. [14) 3.1.1.3 LectinsLectins are proteins capable of binding to carbohydrate moieties of complex carbohydrates without altering the structure of the carbohydrate. They were discovered about 100 years ago when the observation was made that extracts of beans agglutinate red blood cells. Further research showed that many plants contain lectins. They are predominantly found in seeds where they can comprise up to two percent of the total protein. The actual function of lectins in plants is not clearly understood, but they are believed to function as a defense mechanism against a variety of fungal and bacterial pathogens. Deleterious effects were found e.g. against weevil larvae and European corn borer. The mechanism of toxicity is unknown but probably involves the binding of lectin to insect midgut cells and disrupting of cell functions or inhibition of nutrient uptake.[16] The lectin wheat germ agglutinin has been introduced into maize to achieve insect resistance and field trials have been carried out. Hilder et al. transformed tobacco with a snowdrop (Galanthus nivalis) lectin. They reported enhanced resistance of the transgenic tobacco against the sap-sucking peach potato aphid (Myzus persicae). However, no commercially acceptable level of protection was reached.[17] Resistance against insects feeding on phloem sap may be enhanced by using phloem-specific promoters such as the rice sucrose synthase (RSs1) promoter. [15] 3.1.2 Protection against Viral InfectionsViruses cannot be controlled by chemical means. As viruses can cause substantial losses in many crops, they are indirectly controlled by spraying chemicals against virus vectors (mainly insects), by using certified virus-free material, or by eradicating infected plants. As these measures are not overly successful, the engineering of virus resistance into plants is expected to provide a direct and more efficient control.
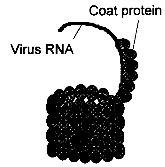
Viruses are genetic elements much smaller than e.g. bacterial cells. They do not have an own matabolism and multiply only inside living host cells using the cell metabolic machinery. Viruses consist of either a DNA or RNA genome surrounded by a coat protein (see fig. 3.1), and occasionally other material. When a virus multiplies, the viral genome is released from the coat into the host cell, where the host metabolism starts produtcion of viral proteins and replication of viral genes, finally resulting in the release of new viruses. About 80% of pathogenic plant viruses have genomes of plus strand single-stranded RNA which may be directly used as mRNA, but also single- or double-stranded DNA viruses cause significatn losses to crops. Most genetic engineering strategies against viruses are based on the introduction of a virus-derived gene sequence into the genome of the host plant. The products of these sequences (mRNA and proteins) interfere with specific stages in the viral infection cycle, such as virus replication or spread, thereby resulting in a virus resistat plant. Resistance mediated by expression of a viral coat protein is the most advanced approach and has already led to resistant varieties which are expected on the market soon. Some other strategies also confer a high degree of resistance, e.g. replicase-mediated protection, but no commercial products have been developed ye using these methods. 3.1.2.1 Coat Protein-Mediated ProtectionBiological Basis The genome of plant viruses encodes a coat protein that protects the viral nucleic acid during the transfer from plant to plant and can determine the specificity of the vector. In coat protein-mediated protection (CPMP), a gene for a viral coat protein is introduced into the genome of the plant. The concept of CPMP has been derived from the observation of cross protection: by preinfection with a mild symptomless virus strain, plants are protected against a related but severely damaging strain. The first report on CPMP was published in 1986, when the Tobacco Mosaic Virus (TMV) coat protein gene was introduced into the genome of tobacco plants. Transgenic plants expressing high levels of coat protein showed a delay in disease development upon infection with TMV.[9] Typically, CPMP is achieved by transforming plants with a functional sense coat protein gene under control of a Cauliflower Mosaic Virus (CaMV) 35S promoter that mediates strong constitutive transcription in many cell types. CPMP works against the virus strain from which the coat protein gene originates and against related viruses. The mechanism of this resistance is not fully understood. The degree of resistance was often correlated with the amount of intact coat protein expressed in transgenic plants and resistance could be overcome by high concentrations of virus inoculum. Several lines of evidence suggest that the coat protein produced. by the plant impedes the uncoating of the viral RNA at the beginning of the infection cycle. In some cases, resistance was found in plants expressing only low levels of coat protein, in plants expressing defective coat proteins, or in plants expressing only coat protein mRNA which was not translated into protein. In these examples, resistance may be mediated by the coat protein mRNA.[18] State of Development So far, CPMP is the only method for genetically engineered resistance which has the potential for agronomic use. CPMP has been demonstrated for more than 20 RNA viruses, but no effect on DNA viruses has yet been reported. Tobacco has been used as a model system to study CPMP, but resistance has now also been engineered into important field crops such as potato, tomato, squash, alfalfa, and has also been extended to cereals.[9] The efficiency of CPMP has been tested in field trials with several plant species (see table 3.2). The first field trial was conducted in 1987 with transgenic tomato plants. After mechanical inoculation with tobacco mosaic virus (TMV), at the time of fruit harvest only 5 % of the transgenic plants expressing the TMV coat protein had developed disease symptoms, compared to 99% of the control plants. The fruit yield was equal to that of non-infected plants.[9] Other trials have shown similarly high degrees of resistance, demonstrating the usefulness of this approach. The US seed company Asgrow has developed a transgenic squash variety resistant to zucchini yellow mosaic virus and watermelon virus II using coat protein genes of the two viruses. Asgrow has obtained market approval and commercial sales are expected soon (see chapter 4.4).
Table 3.3: Field tested crops with a coat protein-mediated protection against
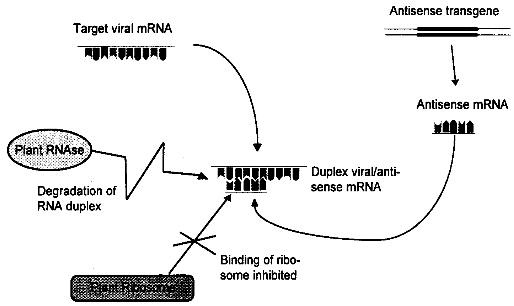
viruses. Abbreviations: AIMV alfalfa mosaic virus, ArMV arabis mosaic virus, BNYVV beet necrotic yellow vein virus, BYDV bailey yellow dwarf virus, BYMV bean yellow mosaic virus, CMV cucumber mosaic virus, MDMV maize dwarf mosaic virus, PLRV potato leafroll virus, PRSV potato ringspot virus, PRV papaya ringspot virus, PVS potato virus S, PVX potato virus X, PVY potato virus Y, RSV rice stripe virus, SMV soybean mosaic virus, TEV tobacco etch virus, TMV tobacco mosaic virus, TRV tobacco rattle virus, TSV tobacco streak virus, TSWV tomato spotted wilt virus, TYLCV tomato yellow leaf curl virus, WMV2 watermelon mosaic virus II, ZYMV zucchini yellow mosaic virus. 3.1.2.2 Replicase-Mediated ProtectionBiological Basis As an alternative to the use of genes encoding structural proteins like coat protein genes, the introduction of sequences coding for non- structural viral proteins has been exploited to create virus resistant plants. There are several reports now describing how expression of a viral RNA-polymerase in transgenic plants leads to resistance. The viral RNA-dependent RNA-polymerase, also called replicase, is used by the virus to generate new RNA copies of the viral RNA genome. The first report was published in 1990, when it was discovered that transgenic tobacco plants expressing part of the replicase gene of a TW were highly resistant to this virus and a closely related strain. The mechanisms conferring resistance are not clear. Resistance was brought about both by replicase genes which coded for functional proteins and by mutated genes which lead to production of non-functional protein or no protein at all. One explanation of resistance is based on the observation that overproduction of one component of the transcription complex (in this case the replicase) can lead to inhibition of transcription. If defective replicase is expressed, resistance may result from the effect of the dysfunctional protein which dominates the functional viral proteins. The efficacy of both proposed mechanisms should increase with enhanced expression of the transgene. However, these explanations are in contrast with several experiments where the highest level of resistance has been observed in the plants with the lowest expression rate of the transgene, an effect which has also been found in coat protein-mediated resistance. In these cases, the resistance may be mediated by mRNA, as proposed by Baulcombe in.[21] State of Development In the last five years, at least eight examples of replicase-mediated resistance in model plants have been published, including resistance against tobacco mosaic virus, pea early browning virus, cucumber mosaic virus, potato viruses X and Y, and cymbidium ringspot virus. Though apparently no field trials have been carried out, the extreme level of resistance observed in some experiments makes it likely that this approach will be useful in the field. A limitation may be the strain specificity of the effect, which could cause problems in genetically heterogenous viral field isolates.[21] 3.1.2.3 Antisense RNA-Mediated ResistanceBiological Basis Antisense RNA is RNA complementary to mRNA of a particular gene. Antisense RNAs were initially recognized in bacteria as naturally occurring mechanisms of gene expression. Antisense strategies are increasingly used as a means of down regulating specific genes. The most famous application to date is the FlavrSavrTM tomato with an extended shelf life which was the first transgenic food available on market. The extended shelf life is achieved by down regulating the polygalacturonase (PG) gene responsible for the softening of the tomato using an antisense gene of the PG gene. Expression of antisense RNA in transgenic plants is achieved by introducing the noncoding strand of a gene together with an appropriate promoter into the plant genome. The mode of action of antisense RNA is not clearly understood. The antisense RNA is thought to form a duplex with the target viral RNA by hybridization. The duplexes formed may stimulate degradation of the mRNA by RNAses or could prevent binding of the ribosome, thereby impeding translation into proteins (see figure 3.2). [22] State of Development Research with antisense genes used for virus resistance has been predominantly conducted with tobacco and potato. The antisense approach was successful in several cases, e.g. in transgenic Russet Burbank potato against potato leafroll virus (PLRV). In this case, sense and antisense constructs against viral coat protein were equally effective. Examples of successful induction of resistance also include tomato golden mosaic virus (TGMV), a single stranded DNA virus which replicates in the nucleus. In several other instances, the protection was less effective, e.g. in tobacco expressing antisense coat protein genes of PVX. [22]
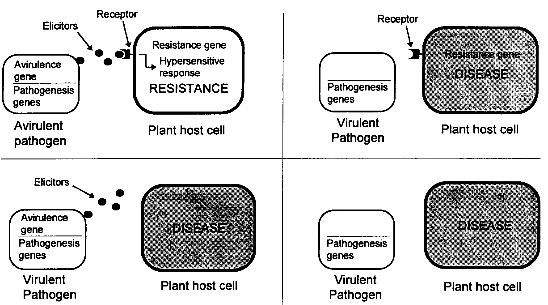
3.1.2.4 Satellite RNA-Mediated Disease AttenuationBiological Basis Satellite RNAs are small extragenomic RNA molecules found in some plant viruses. They cannot replicate individually and depend on a helper virus for replication. They have no sequence similarity with their helper virus, and are thus not directly derived from the helper virus genome. Some satellite RNAs are known to attenuate the symptoms of the helper virus. For example, in China, pepper plants are prophylactically inoculated with a cucumber mosaic virus (CMV) strain which contains satellite RNA. These plants develop only mild symptoms and are protected against infection with a more harmful CW strain not containing the satellite. Transgenic plants expressing satellite RNA were shown to be protected against the severe effects of their helper virus. As a mechanism for satellite RNA-mediated disease attenuation, competition with the viral genomic RNA for a limiting amount of replicase enzyme has been proposed, but other mechanisms seem to contribute to the disease attenuation as well.[9] State of Development The most widely studied satellite RNAs are those associated with cucumber mosaic virus (CMV). Several groups have shown that transgenic plants expressing CMV satellite RNA are tolerant to CMV. Transgenic plants showed almost no symptoms after infection with CMV. Also tobacco plants expressing tobacco ringspot virus (TobRSV) satellite RNA were tolerant to TobRSV.[9] Few researchers envisage field testing of transgenic plants due to the possibilities of producing deleterious satellite RNA (see chapter 4.1.4.2). As mentioned before, cross protection with a CMV strain attenuated by a natural satellite RNA has been shown in the field, not only at a large scale in China, but also at smaller scale in U.S. and Italy. Large-scale tests of transgenic plants expressing satellite RNA are being carried out in China. [23] 3.1.2.5 Defective Interfering RNA/DNA ProtectionDefective interfering (DI) particles are deletion mutants of genomic viral sequences which depend on their parent virus for replication. DI RNAs have frequently been described in animal virus systems but, so far, only rarely in plant virus systems. Like satellite RNAs, DI particles can attenuate the disease symptoms of their parent virus by interfering with its replication, as found for tomato bushy stunt virus (TBSV). As an alternative to the introduction of naturally occurring DI molecules, transformation with artificially constructed Dis have been described. Deletion mutants of several viral RNAs were shown to reduce the replication of the respective parent viruses.[9] 3.1.2.6 Ribozyme-Mediated ProtectionRibozymes are RNA molecules catalyzing RNA cleavage reactions. Mostly, the cleavage site consists of a consensus structure, called the "hammerhead" motive. The nucleotide region directing the catalysis of the cleavage reaction could be separated from the region where the cleavage occurs and the recognition of the target could be modified by changing the nucleotide sequence of the regions flanking the cleavage site. Thus, ribozymes could be designed which cleave viral sequences. Transgenic plants could express a ribozyme which cleaves viral nucleotides. Whether ribozymes will be active in stably transformed plants, and by which expression strategy a sufficient amount of ribozyme can be produced to achieve a reduction in target gene expression remains to be investigated.[9] 3.1.3 Resistance to Fungal Pathogensplants exhibit natural resistance to fungal attack, and disease is the exception rather than the rule. However, the exceptions can be costly and even devastating. Fungal disease remains one of the major factors limiting crop productivity worldwide, with often huge losses set against massive cash inputs for pesticide treatments, exemplified by an estimated cost to the wheat farmers of US$100 million per annum in Europe alone. [24] The total market for fungicides amounts about US$ 5.6 billion world-wide (1991).[1] Hence, there is great interest in the development of novel strategies for protecting crop plants against fungal diseases, to complement traditional plant breeding efforts and to reduce the expense and possible environmental costs of reliance on conventional agrochemical treatments. [24] Although several promising approaches exist, no transgenic varieties with fungal resistance are expected to approach market introduction in the near future. Strategies for obtaining plants resistant to fungi are largely based on natural plant responses to pathogen attack. Therefore, the following chapter starts with a short introduction into the principles of natural plant disease resistance. Although this part deals with resistance to fungi, many of the methods described may also be used to achieve enhanced resistance to other pathogens, e.g. viruses, bacteria, and nematodes. 3.1.3.1 Natural Plant Disease ResistancePlants, like animals, are continually exposed to pathogen attack. Plant- pathogenic organisms are diverse and include viruses, bacteria, fungi, nematodes and protozoa. These pathogens usually live within the plant cell or in the intercellular space. Because plants lack a circulatory system and antibodies, they have evolved a defense system that is distinct from the vertebrate immune system. In contrast to animal cells, each plant cell is capable of defending itself by a combination of different defense reactions. Resistance to a pathogen is often correlated with a "hypersensitive response", induced cell death in the host plant at the site of infection. The hypersensitive response (HR) involves generation of active oxygen species, ion fluxes across membranes, cross-linking and strengthening of plant cell wall, production of antimicrobial compounds (phytoalexins) and induction of pathogenesis- related (PR) proteins such as chitinases and glucanases. Though the mechanisms are unknown, HR is thought to be responsible for the limitation of pathogen growth. The HR is hypothesized to trigger a subsequent response to pathogen attack, referred to as systemic acquired resistance, that acts throughout the plant and not only at the site of infection. Systemic acquired resistance reduces the severity of disease caused by all classes of pathogens, including normally virulent pathogens. [25] Whether a plant is susceptible or resistant to a specific pathogen often depends on its ability to induce HR. Induction of HR follows recognition of specific signal molecules (elicitors) produced by the pathogen. These elicitors are directly or indirectly coded for by so-called avr (for avirulence) genes of the pathogen. The receptors of the plant cell which recognize the elicitors and (indirectly) initiate the HR are coded for by R (for resistance) genes. Therefore, a plant is only protected against pathogens which produce a specific elicitor that is recognized by a specific plant receptor as shown in figure 3.3. [25] In conventional plant breeding, plants with resistance (R) genes against important pathogens have been selected according to their phenotypic traits and the resistance has been introduced into other varieties by hybridization. Unfortunately, resistance of a new variety conferred by a single resistance gene is often overcome after only a few years by the development of a new pathogen race, and the combination of several resistance genes by conventional breeding is even more time- consuming than introduction of a single R gene. Today, the growing understanding of the mechanisms leading to plant disease resistance is increasingly used for plant genetic engineering. Recently, the first R genes have been identified and described in plants. These include R genes from Arabidopsis sp. (resistance to the bacterium Pseudomonas syringae), tobacco (resistance to tobacco mosaic virus and to P. syringae), tomato (resistance to the leaf fungal pathogen Cladosporium fulvum) and flax (resistance to a leaf rust fungal race). [26] The identification of such R genes will not only facilitate the understanding of plant disease resistance, but also makes possible the direct introduction of R genes in different plant varieties using genetic engineering. By combining different R genes responsive to the same pathogen in one plant, resistant varieties with a more durable resistance could be produced. Genes acting at other levels of the plant defense system have already been introduced into plants in order to increase resistance against fungi and other pathogens, as described in the following chapters.
3.1.3.2 Chitinases and GlucanasesChitin and β-1,3-glucans are major structural polysaccharides of the cell walls of many fungi. The breakdown of these components by plant endochitinases and β-1,3-glucanases, both belonging to the group of the pathogenesis-related (PR) proteins of plants which are induced upon pathogen attack, is thought to inhibit fungal growth. Several types of endochitinases and β-1,3-glucanases have been described. A combination of a bean endochitinase gene with the promoter of the CaMV 35S gene conferring high constitutive expression in a wide variety of plant cells was introduced into tobacco. For evaluation, the transgenic tobacco was grown in presence of Rhizoctonia solani, a soilborne fungus that infects numerous plant species. Seedling mortality or the loss of root fresh weight was clearly reduced compared to control plants, depending on the amount of bean chitinase expressed. [27] Similar effects were found with other 35S-chitinase constructs, but in other cases this approach did not reveal a substantial increase of resistance to chitin-containing fungi.[9] Recently, a rice chitinase gene has been introduced into protoplasts of an Indica rice variety using polyethyleneglycol-mediated transformation, and fertile plants have been regenerated. This was one of the first reports of regeneration of the important Indica rice, as these varieties have been particularly difficult to regenerate. Transgenic plants expressing the chitinase showed increased resistance to the fungal sheath blight pathogen Rhizoctonia solani. [28] In another recently published paper, the combined action of chitinase and glucanase was evaluated. The rice RCH10 chitinase and the alfalfa AGLU1 glucanase under control of CaMV 35S promoters were introduced separately into tobacco. The transgenic lines expressing one of the two hydrolytic enzymes were then selfed, and the homozygous progeny lines were crossed with each others. The resulting lines were infected with the fungal pathogen Cercospora nicotianae. Evaluation of disease development showed that the combination of chitinase and glucanase gave the best protection, with only weak symptoms after substantial delay (see figure 3.4). [29]
The finding that combination of genes which alone have only limited effect results in significant protection is promising. Similar results have been found in other experiments (see below). Moreover, combinatorial use of antimicrobial genes may also reduce the breakdown of resistance as a result of pathogen mutation.[29] 3.1.3.3 Ribosome-inactivating ProteinsRibosome-inactivating proteins (RIPs) are plant proteins which inhibit protein synthesis in target cells by cleavage of 28 S RNA. RIPs do not inactivate "self' ribosomes, but show activity towards ribosomes from distantly related species including fungi. Some ribosomes are among the most potent natural toxins, the best known of which is ricin. [24] Purified barley RIP inhibits fungal growth in vitro, and its effect is synergistically enhanced by chitinases and β-1,3-glucanases. These characteristics were used to improve resistance to Rhizoctonia solani in transgenic tobacco. A barley RIP was put under control of the potato wun1 promoter which mediates wound- and pathogen-inducible transcription in the epidermis of leaves, stems and roots from tobacco. Primary transformed regenerates and R1 progeny grew more vigorously in soil inoculated with Rhizoctonia solani than non-transformed control plants. [9] 3.1.3.4 PhytoalexinsIn several host/pathogen systems a correlation has been found between the concentration of phytoalexins (antimicrobial compounds synthesized by plants) and resistance to specific pathogens. In Vitis vinifera and Picea sitchensis, stilbenes are involved in protection against fungal challenge. Most plants, for example tobacco, contain the precursor for the formation of stilbenes but lack the enzyme stilbene synthase. After transfer of a stilbene synthase gene from grapevine to tobacco, transgenic plants expressed the foreign gene in response to treatment with fungal elicitor or infection with the broad range fungal pathogen Botrytis cinerea. This work supports the assumption that at least in some host/pathogen systems the synthesis of phytoalexins plays a crucial role for the defense of the host plant. [9] 3.1.3.5 Other Antifungal ProteinsA large number of other proteins with inhibitory effects on the growth of fungi in vitro'have been identified from plants. Among these are osmotin, a vacuolar protein from tobacco produced in response to salt stress, and small, basic, cysteine-rich proteins of various origins. Also some microorganisms produce such antifungal peptides. [9] These proteins will be tested - alone or in combination - for their effect against fungi in transgenic plants and at least some of them are likely to enhance resistance in transgenic crop plants. 3.1.3.6 Hypersensitive Cell DeathIn the past, crossing-in of race-specific resistance was frequently applied in breeding programs to protect novel varieties against fungal and other diseases, as mentioned before. However, this procedure provides protection only against a limited number of pathogen races and large- scale growth of new varieties led to the rapid selection of new virulent races and epidemic spread in monocultures. Of course, the same problems could also arise if transgenic plants carrying race- specific resistance genes from other cultivars or other species were grown in the field. Different strategies to obtain durable resistance to a wide range of pathogens using artificial generation of programmed cell death are being pursued. One of the most advanced was proposed by Strittmatter et al... [9] A promoter fragment of the potato prp11 gene, which mediates rapid and local transcriptional activation selectively after fungal infection, has been combined with the barnase gene from Bacillus amyloliquefaciens encoding a highly cytotoxic RNase. The resulting chimeric construct was transformed into potato. Rapid synthesis of this RNase in the vicinity of infection sites should initiate necrosis of host cells during early stages of compatible interactions and, therefore, restrict the growth and propagation of biotrophic pathogenic fungi also in this type of interaction, analogous to the naturally occurring hypersensitive cell death in incompatible interactions. Transgenic plants were simultaneously transformed with a chimeric construct which constitutively produces barstar the specific barnase inhibitor from Bacillus amyloliquefaciens. The authors expect, that only in the close vicinity of infection sites the level of cytotoxic barnase will exceed the inhibitor barstar, resulting in strictly localized cell death after infection. 3.1.3.7 Systemic Acquired ResistanceAs mentioned in chapter 3.1.3.1, a hypersensitive response is often followed by a subsequent response called systemic acquired resistance (SAR). SAR acts throughout the plant and protects it against a wide range of pathogens. Establishment of SAR is correlated with the coordinate expression of several groups of genes, so-called pathogenesis-related (PR) proteins. PR-1a is the most highly expressed of these proteins. The gene coding for PR-1a was brought under control of an enhanced 35S promoter and transformed into tobacco. Transgenic tobacco lines showed constitutive high-level expression of the PR-1a protein. This resulted in significant tolerance to infection by the two fungal pathogens Peronospora tabacina and Phytophthora parasitica var. nicotianae. [30] Although the function of PR-1a protein and other PR proteins is not known yet, high-level expression of such proteins may be useful in creating crops with increased resistance to fungal and other diseases. 3.1.4 Resistance to Bacterial PathogensBacterial diseases are a persistent problem, with no satisfactory alternative to copper sulfate treatment.[24] As these treatments cause environmental problems, there is a great interest in the development of novel strategies. While scientific progress has been im- pressive, examples of agronomically significant resistance to plant-pathogenic bacteria are still scarce.[4] 3.1.4.1 Lysozyme-Mediated ResistanceLysozymes, which catalyze the hydrolytic cleavage of bacterial cell wall murein, have been detected in many plant species. As the biochemical and molecular characterization of plant lysozymes and the corresponding genes is still in its infancy, lysozyme genes from other sources have been used for genetic engineering. The expression of lysozyme genes from hen egg white or bacteriophage T4 was achieved in tobacco and potato. One group found an increased resistance of tuber slices from transgenic potato lines to the pathogenic soil bacterium Erwinia carotovora spp. atroseptica.[9] 3.1.4.2 CecropinsCecropins are a family of proteins which form an important component of the immune response of diverse insects. They exhibit antibacterial activity against several Gram-positive and Gram-negative bacteria in vitro, apparently by forming ion channels in the bacterial membrane leading to leakage of cell components and ultimately cell death. [31] A gene coding for cecropin B from the giant silkmoth Hyalophora cecropia was introduced into tobacco Although considerable amounts of cecropin-mRNA were present in transgenic plants, no cecropin B protein could be detected. As rapid degradation of cecropin B was shown in crude plant extracts, the undetectable level of cecropin B in transgenic tobacco is thought to result from degradation of the protein by plant-endogenous proteases. Transgenic plants therefore showed no resistance against the bacteria Pseudomonas solanacearum and Pseudomonas syringae, which are highly susceptible to cecropin B in Vitro.[31] In another approach, substitution analogs of cecropin were produced. In these cecropin analogs, the amino acid sequence was changed but hydrophobic properties and charge density was conserved. One of these substitution analog called Shiva-I with 46 % homology in amino acid sequence was found to have a more potent lytic activity than another cecropin analog with 95 % homology. The gene sequence for Shiva-I was introduced into tobacco. Transgenic tobacco seedlings exhibited delayed wilt symptoms, reduced disease severity and reduced mortality after infection with a highly virulent strain of Pseudomonas solanacearum. [32] 3.1.4.3 Activation of Plant Defense MechanismsPhytoalexins are low molecular weight antimicrobial compounds which are synthesized in plants in response to pathogen attack (see chapter 3.1.3.1). The synthesis of phytoalexins is in most cases based on complex biosynthetic pathways which are not accessible for genetic engineering yet. An exception are thionins, small cysteine-rich polypeptides which were found in endosperm and leaves of cereals. They were shown to exert antimicrobial activity in vitro. One group obtained high level synthesis of functional thionin in transgenic tobacco leaves transformed with a barley a-thionin gene under control of the CaMV 35S promoter. After inoculation with Pseudomonas syringae pv. tabaci or Pseudomonas syringae pv. syringae, the number of necrotic lesions and the severity of disease symptoms were reduced in leaves of transgenic plants compared to control plants. The level of resistance coincided with the level of thionin expression. [9] 3.1.4.4 Detoxification of Pathotoxins and Alteration of Target EnzymesPathogens that produce phytotoxins causing disease symptoms usually have the capacity to metabolize, i.e. detoxify these compounds.[4] For instance, the dipeptide tabtoxin is thought to be responsible for chlorosis during wildfire disease on tobacco caused by Pseudomonas syringae pv. tabaci. The pathogen protects itself against the toxin by expression of the tabtoxin resistance gene, ttr, which encodes an enzyme that acetylates tabtoxin. Transgenic tobacco plants constitutively expressing the ttr gene under control of the CaMV 35S promoter did not produce the chlorotic halo typical for wildfire disease on non-transgenic plants.[9] In a comparable strategy, protection against the damaging effects of a bacterial toxin was obtained by transformation of plants with a pathogen-derived enzyme which is unsusceptible to the toxin. Transgenic tobacco plants harboring a gene construct derived from Pseudomonas syringae pv. phaseolicola expressed an ornithine carbomyl transferase which is insensitive to the phaseolotoxin produced by Pseudomonas syringae pv. phaseolicola. Progeny of these plants displayed a reduction in disease symptoms compared to control plants.[9] 3.2 Protection Mediated by Transgenic MicroorganismsIn contrast to the plant protection approaches based on transgenic plants discussed above, the following chapters describe methods which make use of genetically altered viruses and bacteria, mostly to achieve plant protection against insect pests. Mainly insect-pathogenic baculoviruses and bacteria other than Bacillus thuringiensis expressing Bt toxin have been developed. 3.2.1 Transgenic BaculovirusesBiological Basis Baculoviruses are pathogens of insects that infect predominantly holometabolous insects. Almost all Lepidopterans, which include many of the world's most serious pests, are susceptible to at least one of the more than 500 baculovirus species. The baculoviruses were first studied for use against forest pests in the 1930s and were used until the advent of chemical pesticides in 1960. The virus particles are generally applied on the foliage by spray. The viruses have no contact action and must therefore be ingested by larvae, thus good coverage of plants is required. Of particular interest for insect pest management is Autographa californica nuclear polyhedrosis virus (AcNPV), originally isolated from the alfalfa looper, and the Bombyx mori nuclear polyhedrosis virus (BmNPV).2 The DNA of a baculovirus is enclosed within a so-called nucleocapsid, about 50 by 300 nm in size, which again is enclosed within an occlusion body composed of the protein polyhedrin. These occlusion bodies, each of them containing several nucleocapsids, are referred to as polyhedra. The polyhedra protect the virions in the environment. After ingestion of the polyhedra by an insect, the polyhedrin is dissolved and the nucleocapsids fuse to the midgut cells and migrate to the nucleus, where replication takes place. The produced progeny nucleocapsids spread and infect other cells and tissues, resulting in infection throughout the insect. Nucleocapsids may then be enclosed in new polyhedra synthesized in the nucleus. Before the death of the host, the polyhedra can account for up to 30% of the insect dry weight. The insect may continue to feed for several weeks before death from viral infection. The cadaver filled with virus ruptures easily, releasing millions of polyhedra onto surrounding foliage and soil. [33] The fact that baculoviruses take weeks for killing their hosts is one of the most important disadvantages associated with the use of baculoviruses as insecticides. It has restricted their use to crops capable of sustaining some damage without too much economic loss. In order to decrease the killing time of the baculoviruses and the crop damage, genes of different origins have been introduced into AcNPV, including genes encoding insect hormones, insect-specific toxins and insect enzymes.[33] State of development Field trials with genetically engineered baculoviruses were carried out in England in 1986, 1987 and 1988. The trials examined the persistence of genetically altered AcNPV and were among the first deliberate releases of transgenic organisms.[34] The first recombinant baculovirus with an enhanced insecticidal effect was developed in 1989. The gene of a diuretic hormone which plays an important role in the regulation of the water balance in the insect Manduca sexta (tobacco hornworm) was incorporated into Bombyx mori nuclear polyhedrosis virus (BmNPV). Bombyx mori larvae infected with the transgenic virus died 20% more quickly than larvae infected with wild-type BmNPV. The expression of a toxin of the North African scorpion Androctonus australis in AcNPV resulted in a 50 % reduction of feeding damage, and lethal time was reduced by 25 % compared to wild-type viruses. A toxin from the straw itch mite Pyemotes tritici also introduced into AcNPV to enhance its insecticidal activity reduced time to kill by 30-40%. The titre of juvenile hormone (JH) in haemolymph determines the course of larval development. A reduction in JH titre controlled by juvenile hormone esterase (JHE) leads to cessation of feeding before a moult and initiates metamorphosis. By expression of modified JHEs in AcNPV, insecticidal viruses with lethal times more than 30 % lower than with wild-type viruses were generated [33] Currently, no commercial application of genetically engineered insect viruses is in sight, but their use is thought to have considerable potential. The company American Cyanamid e.g. will conduct field trials with transgenic baculoviruses expressing scorpion toxin in the USA. [35] 2 The viruses are sometimes wrongly called "Autographa californica' or "Bombyx mori" only. These are the scientific names of the insects from which the viruses were first isolated. 3.2.2 Transgenic Bacteria3.2.2.1 Inactivated Pseudomonas Fluorescens for Bt Toxin DeliveryBiological Basis In this approach, a non-pathogenic strain of Pseudomonas fluorescens bacteria is used as host for the Bacillus thuringiensis (Bt) toxin (see chapter 3.1.1.1). In an attempt to improve the foliar persistence of Bt insecticidal activity, a delivery system based on recombinant P. fluorescens expressing Bt toxin which are killed by chemicals prior to field release, has been developed by Mycogen Corporation (San Diego, California). When transformed cells are grown in culture, a typical Bt δ-endotoxin crystal is formed within the cell. While still in the fermentation tank, the P. fluorescens cells are chemically treated in order to kill the cells and cause the cell wall to become more rigid through cross-linking of cell wall components. The cell wall of the dead bacteria now serves as a protective microcapsule for the enclosed δ-endotoxin. This Bt toxin delivery system has been called CellCap ® by Mycogen. [36] State of Development Since no living transgenic microorganisms are released into the environment, this approach has the advantage of relative freedom from environmental and safety concerns associated with field releases of living transgenic organisms. The encapsulated Mycogen product with a lepidopteran active δ-endotoxin was the first recombinant product approved for outdoor testing. Field experiments conducted during 1988 suggested that a foliar application of this engineered microbe protected cabbage from lepidopteran pests for 7 days, whereas insecticidal activity from traditional Bt sprays dissipated by 3 days post-application. This resulted in better insect control, and therefore higher yields. Similar tests have been conducted with engineered microbes targeted against the Colorado potato beetle on potatoes. It was shown that the protected δ-endotoxin could be applied at two thirds the toxin rate and still maintain higher levels of insect control than the full rate delivered by Bacillus thuringiensis. U. S. federal registrations for the first two genetically engineered bioinsecticides using Mycogen's CellCape® encapsulation system, MVP® (lepidopteran-active biotoxin) and M-TrakTM (coleopteran-active biotoxin) were received in 1991 from the Environmental Protection Agency. Commercial sales began that year. [36] 3.2.2.2 Bacterial Endophytes Expressing Bacillus Thuringiensis ToxinsBiological Basis The term endophyte is used for plant-associated microorganisms that live within their host plants. Genetic engineering of nonpathogenic endophytes presents an opportunity for delivery of biopesticides in the plant, i. e. the production of the pesticidal agent within host plant tissues without direct genetic manipulation of the host plant. Most approaches make use of Bacillus thuringiensis (Bt) toxin. The major drawback in using Bacillus thuringiensis directly as a biopesticide lies in the fast degradation of the Bt toxin after spraying the bacteria with the Bt toxin crystal outside the spore (see chapter 3.1.1.). A systemic delivery system using bacteria living within the plant would protect the Bt toxin from degradation, providing consistent levels of protection throughout the growing season by the presence of the active ingredient produced by the endophyte. Areas difficult to reach with chemical sprays (e.g., the inside of stalks) would be better protected from plant-feeding insects by endophytes expressing Bt toxin. A number of plant-associated bacteria have been transformed with Bt toxin genes, including Pseudomonas fluorescens, Pseudomonas cepacia, Rhizobium meliloti and Rhizobium leguminosarum. [8] In the following, the use of transgenic Clavibacter as biopesticide will be described in more detail, as this application is one of the most advanced in this field. [37] State of Development The Gram-negative bacterium Clavibacter xyli ssp. cynodontis (Cxc) is a common endophyte living in the xylem of Bermudagrass (Cynodon dactylon) and has been found in this host in the United States, Japan, Taiwan, and France. It has also been found in a few weed species, and it can colonize maize, rice, sorghum, oats, white millet and sudangrass through artificial inoculation. The gene coding for the CrylA(c) protein from Bt ssp kurstaki was inserted into the chromosome of a wild-type Cxc. Unlike Bt, transgenic Cxc do not release the Bt toxin. Instead, the cell must be digested by the insect in order to release the active ingredient in the gut. The observed effects are similar to those observed in larvae ingesting Bt (see chapter 3.1.1.). Field studies have demonstrated that a wide range of commercial field and sweet maize hybrids can be inoculated successfully, with 80-100 % of the inoculated seeds producing vigorous, endophyte colonized plants. The European corn borer (ECB), Ostrinia nubilalis, is cosmopolitan in its distribution and a significant pest in maize. Maize plants inoculated with recombinant Cxc; and artificially infested with ECB larvae sustained up to 80 % less damage than plants inoculated with either wild type Cxc or uninoculated plants. C= was the first recombinant microorganism expressing Bt toxin which has been approved for field testing [37]
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||




 antibodies on hand
antibodies on hand 